Abstract

Background: Activating somatic mutations of STAT3 are one of the most frequently recognized genetic alterations in T-cell lymphoma including T-cell large granular lymphocytic leukemia (T-LGLL). T-LGLL is associated with certain proportions of patients (pts) with acquired aplastic anemia (AA) and/or myelodysplastic syndromes (MDS). Pure red cell aplasia (PRCA), especially acquired PRCA, is a syndrome defined by an anemia with marked reduction or absence of erythroid production associated with immunological mechanisms. PRCA is classified into primary and secondary types including T-LGLL. Previous reports have described that T-LGLL with STAT3 mutations was closely associated with PRCA. However, it is yet to be clear whether STAT3-mutated T cells would be involved in the pathogenesis of other cytopenias, such as AA, MDS, primary PRCA or secondary PRCA without (w/o) T-LGLL. In this study, we analyzed the profiles of STAT3 gene mutations in bone marrow failure syndrome (BMFS)s including AA/MDS and PRCA.
Methods: Peripheral blood samples were collected from 94 patients with BMFSs, including AA, MDS, AA/PNH, and PRCA with or w/o T-LGLL. When possible, T-cell receptor (TCR) Vβ repertoire of the patients was determined by flow cytometry. DNA was extracted from mononuclear cells (MNC), and STAT3 Y640F and D661Y, two hot spot mutations, were detected with sensitive allele specific-PCR (AsPCR) methods. Then we performed deep amplicon sequencing analysis of STAT3 in PRCA pts. CD8+ T cells were sorted if possible, and also subjected to the analyses.
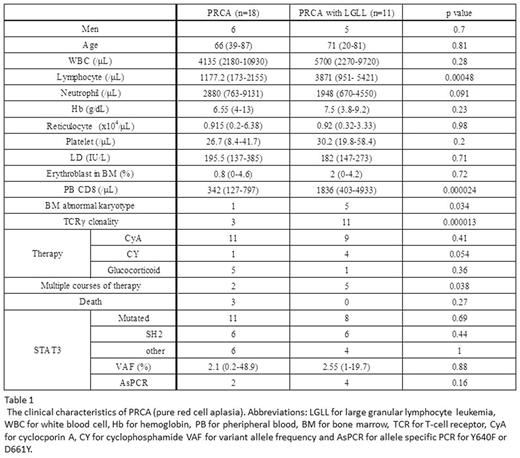
Results: The recruited pts included 38 AA, 20 MDS, 7 AA/PNH, 11 PRCA with T-LGLL, and 18 PRCA w/o T-LGLL. Among PRCA pts w/o apparent T-LGLL, 10 had primary PRCA and 8 had secondary PRCA including 2 with thymoma. The clinical characteristics of PRCA are shown in Table 1. TCR Vβ repertoire analysis were performed in 8 PRCA w/o T-LGLL and 5 PRCA with T-LGLL. All pts with PRCA associated with T-LGLL showed restricted Vβ repertoire. On the other hand, only 1pt showed monoclonal Vβ pattern in PRCA w/o TLGLL. AsPCR did not detect Y604F or D661Y mutations in any of 65 pts with AA, MDS, or AA/PNH. In contrast, 2 pts with PRCA and 4 pts with PRCA-T-LGLL were positive for either of the mutations, suggesting STAT3 mutations may be unique to PRCA pts. Further analysis with deep amplicon analysis in the 29 PRCA pts revealed STAT3 mutations in 11 pts (61%) of PRCA w/o T-LGLL, which included 6 pts with primary PRCA and 5 pts with secondary PRCA associated with autoimmune diseases (n=2), 2 thymoma (n=2) and thymic cancer (n=1). The median variant allele frequency of STAT3 mutations was 2.3% (0.2-48.9). N538Y and I653V were most frequent mutations in PRCA w/o T-LGLL. Eight pts (73%) with T-LGLL-associated PRCA were also positive for STAT3 mutation. Thirty-nine % and 70% of STAT3 mutation were found in SH2 domain in PRCA w/o T-LGLL and PRCA with T-LGL, respectively. There was no difference in clinical features between pts with and w/o STAT3 mutations.
Discussion: The high frequency of STAT3 mutations in PRCA pts regardless of etiology and the absence of the mutation in pts with AA/MDS suggest that STAT3 mutations may serve as a molecular marker of PRCA. The discordant results with regard to AA/MDS pts from a previous report which detected STAT3 mutations in 4.9% of AA/MDS patients by AsPCR methods (Jerez et al. BLOOD 2013) may be due to the difference in ethnic backgrounds of the pts studied. Given very low frequencies of STAT3 mutations in AA and MDS that were revealed by several previous studies, JAK/STAT signaling systems in the axis of STAT3 mutations may have unique roles in the pathogenesis of PRCA.
Conclusion: STAT3 mutations are frequently detected in pts with PRCA even in pts w/o T-LGLL and the JAK/STAT pathway may therefore be therapeutic targets of PRCA.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal